Common sources of 5′end soft clipping and mismatches in 3′ end seq. (a)... | Download Scientific Diagram
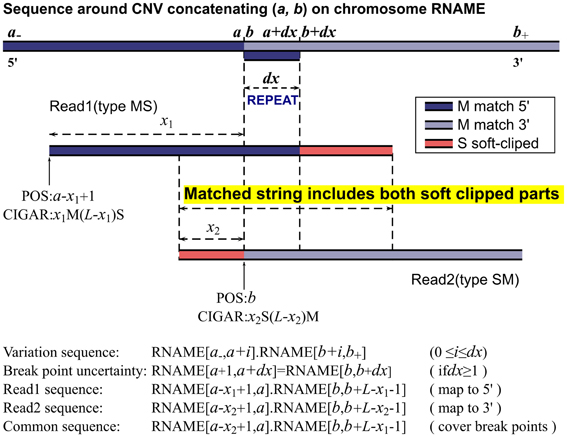
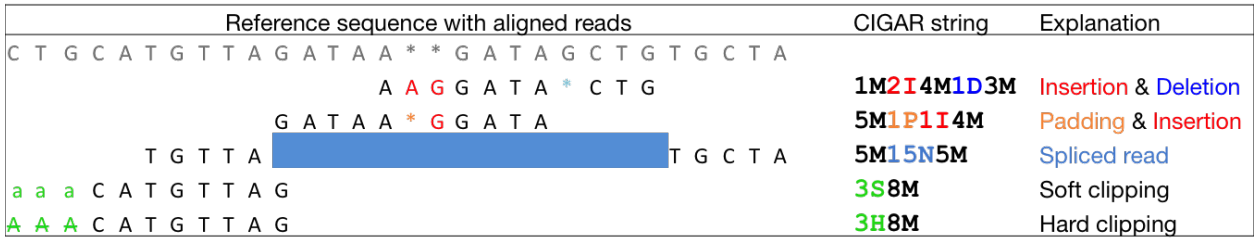
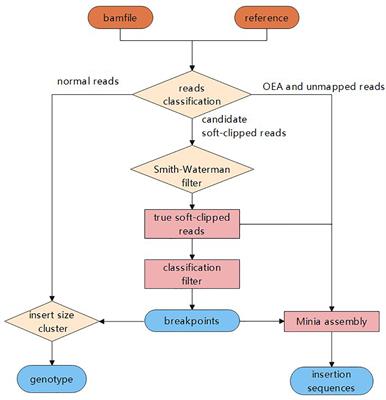
Frontiers | MATCHCLIP: locate precise breakpoints for copy number variation using CIGAR string by matching soft clipped reads

Soft-clipped reads vs. improperly aligned reads at a structural variant... | Download Scientific Diagram

Accurate Detection and Quantification of FLT3 Internal Tandem Duplications in Clinical Hybrid Capture Next-Generation Sequencing Data - ScienceDirect
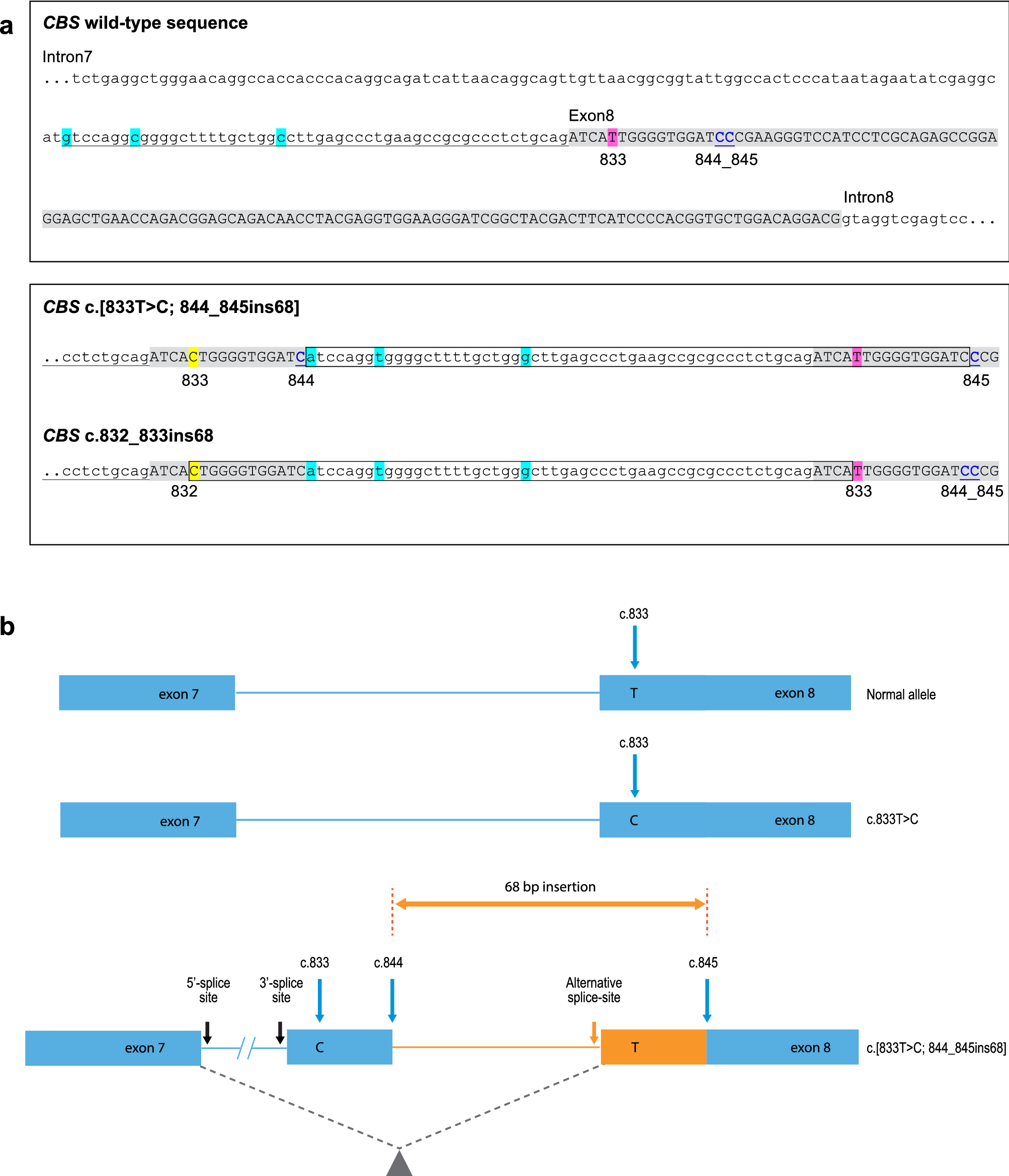
A customized scaffolds approach for the detection and phasing of complex variants by next-generation sequencing | Scientific Reports
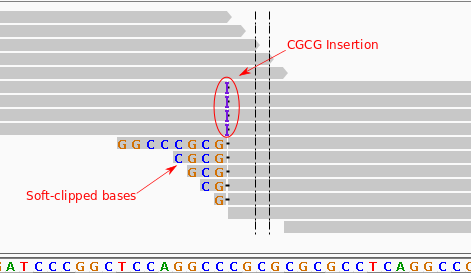
sequence alignment - What is local realignment and what is the problem it solves? - Bioinformatics Stack Exchange
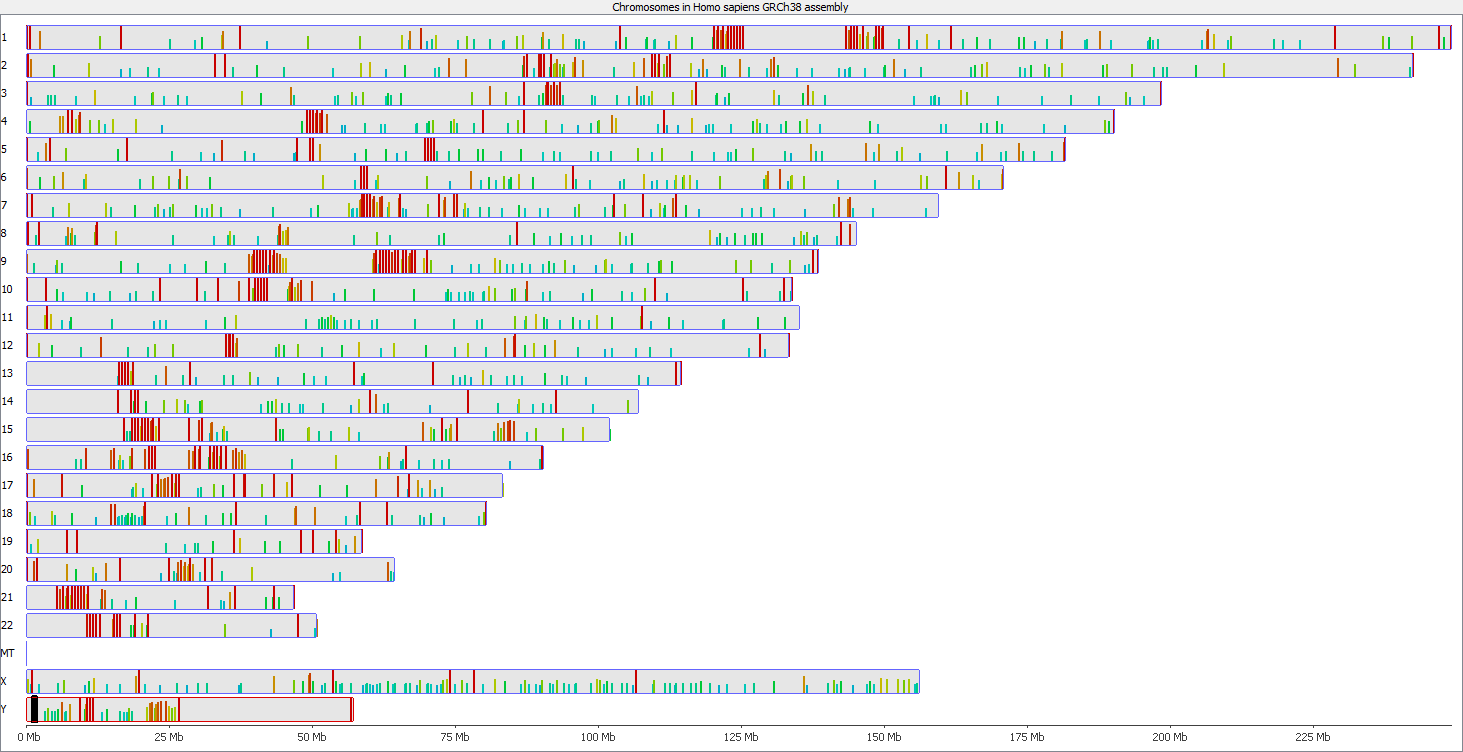
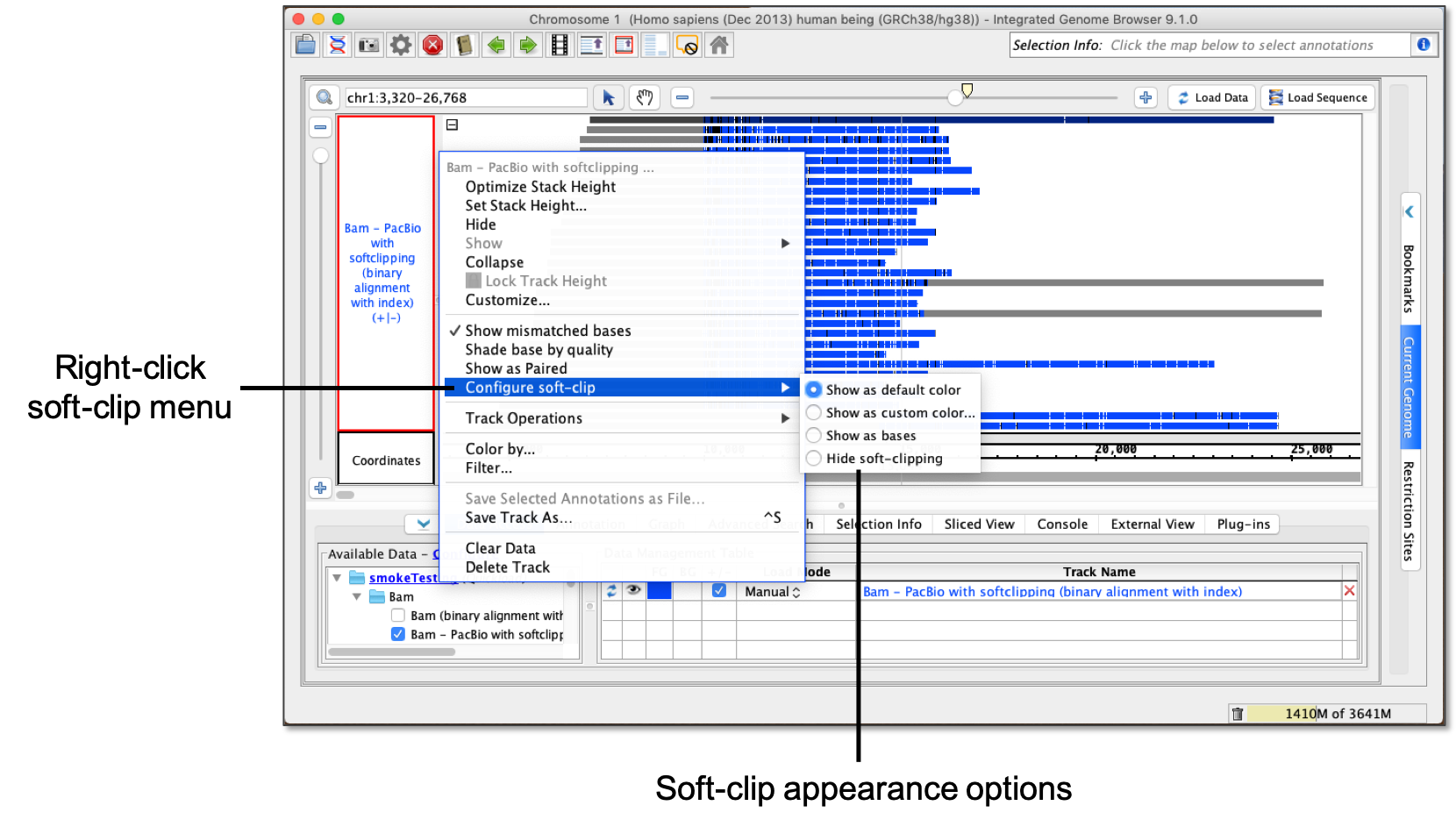
QC Fail Sequencing » Soft-clipping of reads may add potentially unwanted alignments to repetitive regions

VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. - Abstract - Europe PMC
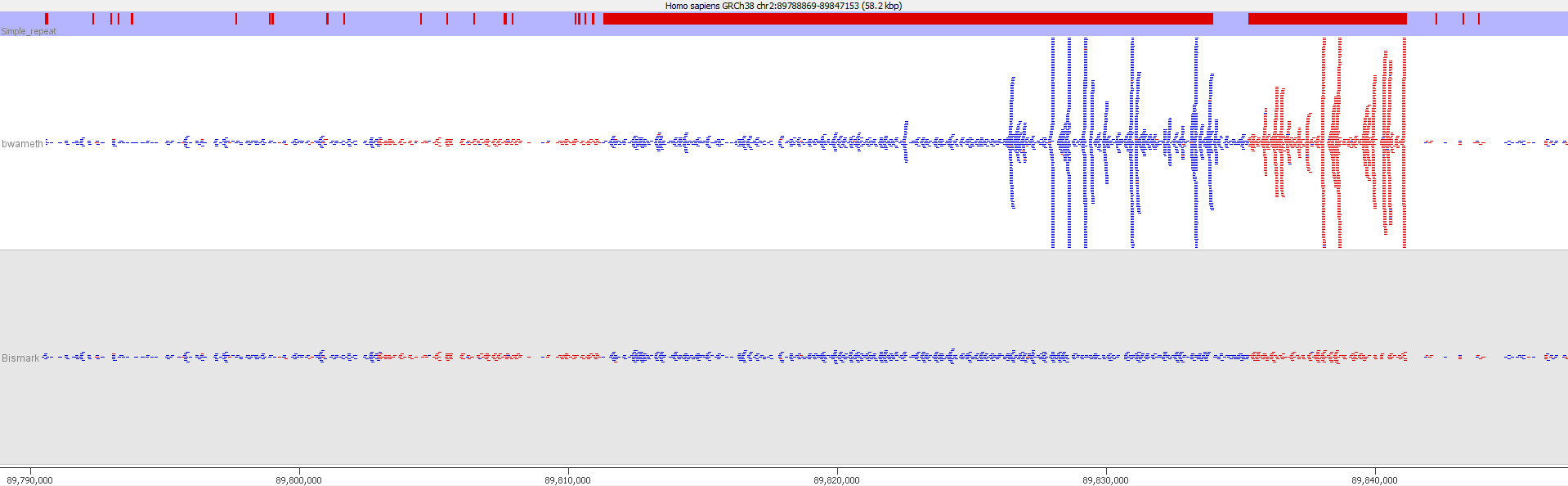
QC Fail Sequencing » Soft-clipping of reads may add potentially unwanted alignments to repetitive regions
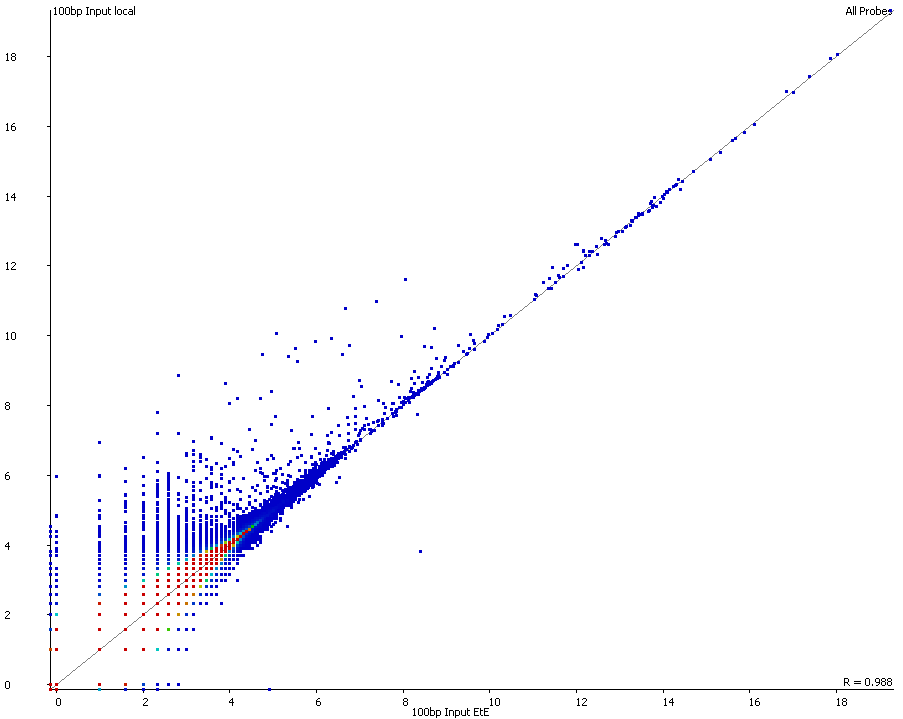
QC Fail Sequencing » Soft-clipping of reads may add potentially unwanted alignments to repetitive regions

Soft-clipped reads vs. improperly aligned reads at a structural variant... | Download Scientific Diagram
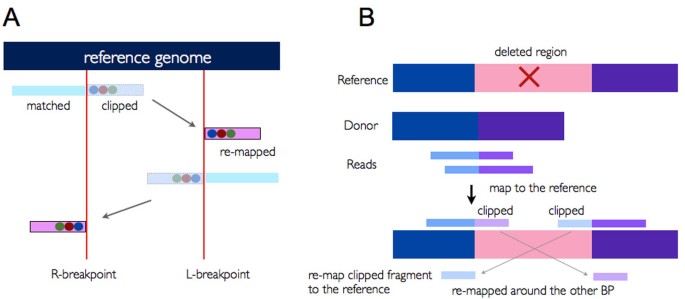
ClipCrop: a tool for detecting structural variations with single-base resolution using soft-clipping information | BMC Bioinformatics | Full Text
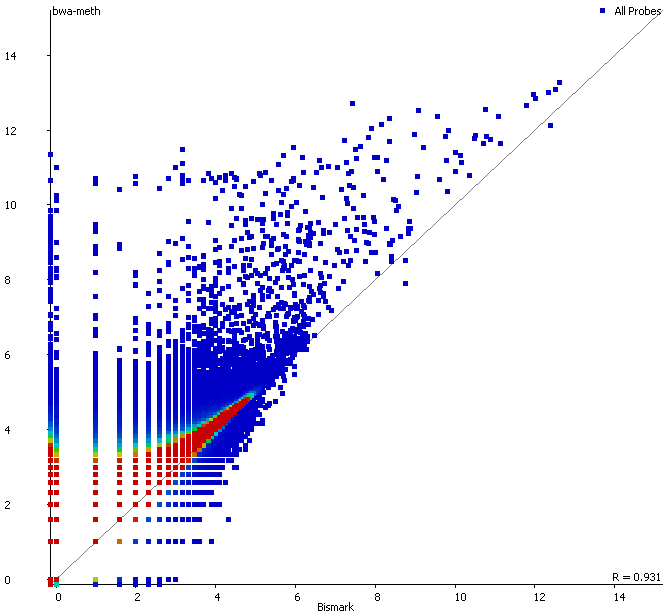
QC Fail Sequencing » Soft-clipping of reads may add potentially unwanted alignments to repetitive regions
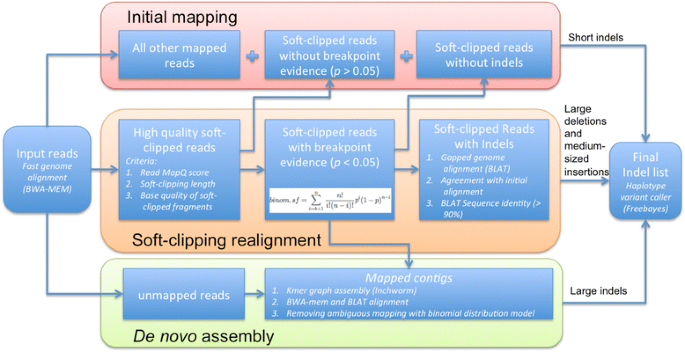
ScanIndel: a hybrid framework for indel detection via gapped alignment, split reads and de novo assembly | Genome Medicine | Full Text

eKLIPse: a sensitive tool for the detection and quantification of mitochondrial DNA deletions from next-generation sequencing data - ScienceDirect

IMSindel: An accurate intermediate-size indel detection tool incorporating de novo assembly and gapped global-local alignment with split read analysis | Scientific Reports
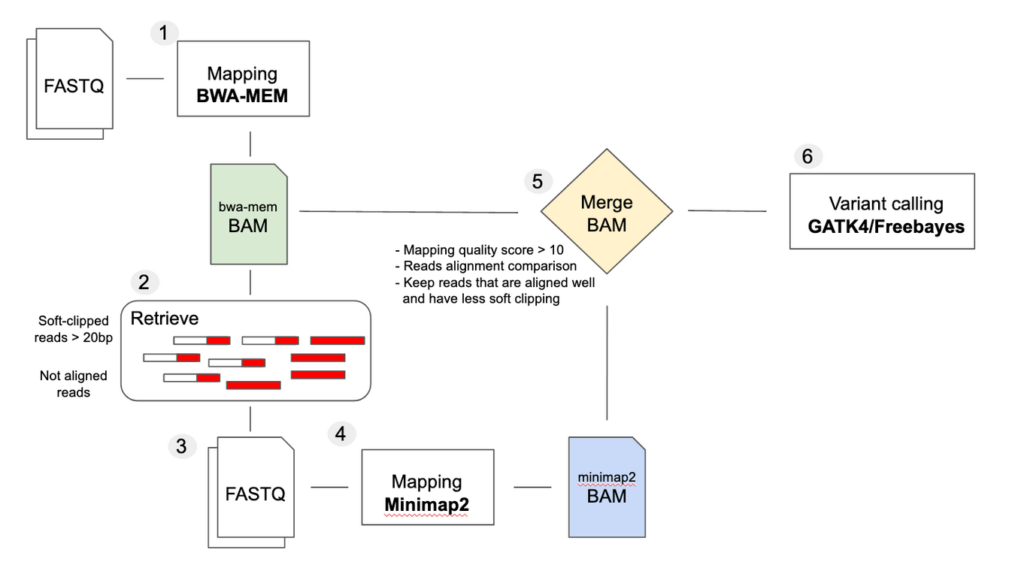
Challenges and importance of mid-sized deletion identification for genomic medicine - SeqOne Genomics

Common sources of 5′end soft clipping and mismatches in 3′ end seq. (a)... | Download Scientific Diagram
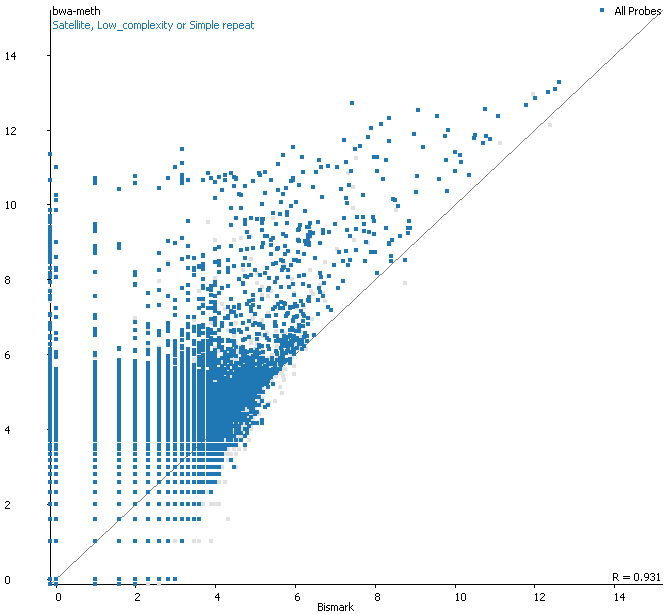
QC Fail Sequencing » Soft-clipping of reads may add potentially unwanted alignments to repetitive regions
![PDF] Socrates: identification of genomic rearrangements in tumour genomes by re-aligning soft clipped reads | Semantic Scholar PDF] Socrates: identification of genomic rearrangements in tumour genomes by re-aligning soft clipped reads | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/16b34a01c549bc1b12b6c5a17e85e1db117f26e3/2-Figure1-1.png)